Exploring Semantic Segmentation with UNets
Introduction
UNets are deep neural networks for segmentation tasks. There have been several segmentation models developed over the years, including VGG and FCN that produce state of the art results on large datasets. This article explores some of the elements of a U-Net and trains the network on a subset of the Pets dataset to perform semantic segmentation. Designed for biomedical data, U-Nets excel at precise segmentation while preserving image size on relatively small datasets.
Model
Like many previous segmentation models, UNets essentially map image inputs to segmentation masks by compressing inputs through a latent space. However, in contrast to previous approaches, this model employs skip connections to deliver additional information to intermediate layers. This gives the model architecture the distinctive ‘U’ shape from which it derives its name. A diagram of the model can be seen below.
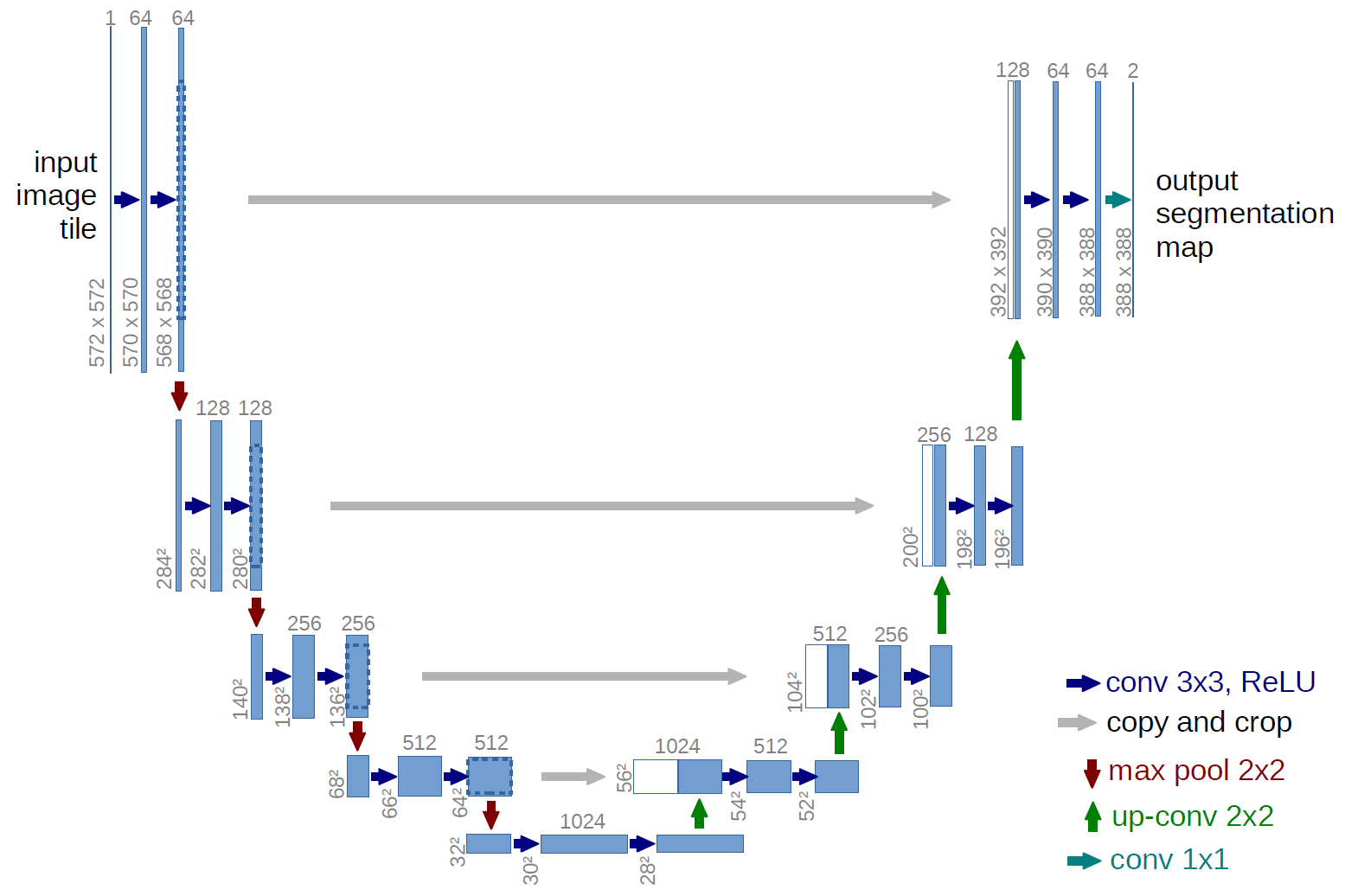
Data Preparation
I trained the model on the pets data. For further simplicity, I filtered the dataset to only the dogs which contains about 5000 images. The dogs dataset also contained an assortment of dog breeds, various different positions (sitting, standing) and different scales. These variations in the dataset still provide some challenge to the model in learning to segment dogs. An example of the data is given below.
I trained the model on the Pets dataset, specifically focusing on the subset of dog images, encompassing approximately 5000 images. The dog dataset featured a diverse range of dog breeds, in various positions (sitting, standing), and scales. These variations within the dataset help to challenge the network in effectively segmenting any dog in any scale or position. Finally, the input images are resized to 150x150 due to resource constraints and simplicity. An example of the data is provided below

Loss Function
For this network, I employed a loss function that combines two distinct loss functions. Based on the number of classes, either binary cross-entropy or cross-entropy can be utilized. Because this model only segments dogs and one can expect only a single target to segment in an image, we can use the binary cross-entropy loss function to help learn the segmentation mask. Additionally, I augmented the loss function by incorporating an additional dice loss. This loss utilizes the Dice coefficient, which is a measure of how well the predicted mask aligns with the true mask. The pseudo description of the loss function is given below.
\[Loss(true\_mask, pred\_mask) = BCE(true\_mask, pred\_mask) + DiceLoss(true\_mask, pred\_mask)\]Results
I’ve skipped over the code for brevity but it can be found on github. To keep it simple, the model was trained for only two epochs. After these epochs, the model achieved a dice coefficient score of approximately 0.858 on the testing dataset (1 being perfect). Checkout the model’s segmentation results below.

The figure above demonstrates the model’s performance on five outputs. The true masks are displayed in the center, while the predicted masks are positioned in the rightmost column. These results are remarkably good considering the brief training period and modest dataset size. Further epochs of training and a larger dataset (augmentation) is likely to significantly improve the results seen above.